12.1. Introduction¶
This section describes some of the general characteristics of RMG’s databases.
12.1.1. Group Definitions¶
The main section in many of RMG’s databases are the ‘group’ definitions. Groups are adjacency lists that describe structures around the reacting atoms. Between the adjacency list’s index number and atom type, a starred number is inserted if the atom is a reacting atom.
Because groups typically do not describe entire molecules, atoms may appear to be lacking full valency. When this occurs, the omitted bonds are allowed to be anything. An example of a primary carbon group from H-Abstraction is shown below. The adjacency list defined on the left matches any of the three drawn structures on the right (the numbers correspond to the index from the adjacency list).
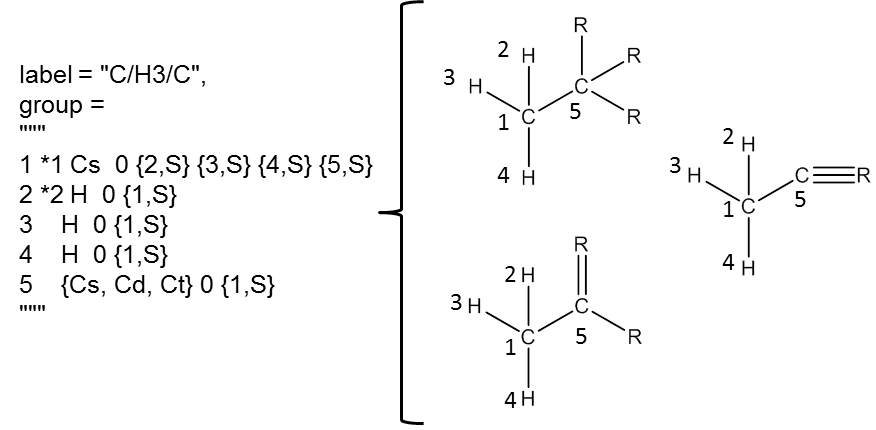
Atom types describe atoms in group definitions. The table below shows all atoms types in RMG.
Atom Type |
Chemical Element |
Localized electronic structure |
|---|---|---|
R |
Any |
No requirements |
R!H |
Any except hydrogen |
No requirements |
H |
Hydrogen |
No requirements |
C |
Carbon |
No requirements |
Ca |
Carbon |
Atomic carbon with two lone pairs and no bonds |
Cs |
Carbon |
Up to four single bonds |
Csc |
Carbon |
Up to three single bonds, charged +1 |
Cd |
Carbon |
One double bond (to any atom other than O or S), up to two single bonds |
Cdc |
Carbon |
One double bond, up to one single bond, charged +1 |
CO |
Carbon |
One double bond to an oxygen atom, up to two single bonds |
CS |
Carbon |
One double bond to an sulfur atom, up to two single bonds |
Cdd |
Carbon |
Two double bonds |
Ct |
Carbon |
One triple bond, up to one single bond |
Cb |
Carbon |
Two benzene bonds, up tp one single bond |
Cbf |
Carbon |
Three benzene bonds (fused aromatics) |
C2s |
Carbon |
One lone pair, up to two single bonds |
C2sc |
Carbon |
One lone pair, up to three single bonds, charged -1 |
C2d |
Carbon |
One lone pair, one double bond |
C2dc |
Carbon |
One lone pair, one double bond, up to one single bond, charge -1 |
C2tc |
Carbon |
One lone pair, one triple bond, charged -1 |
N |
Nitrogen |
No requirements |
N0sc |
Nitrogen |
Three lone pairs, up to one single bond, charged -2 |
N1s |
Nitrogen |
Two lone pairs, up to one single bond |
N1sc |
Nitrogen |
Two lone pairs, up to two single bonds, charged -1 |
N1dc |
Nitrogen |
Two lone pairs, one double bond, charged -1 |
N3s |
Nitrogen |
One lone pair, up to three single bonds |
N3d |
Nitrogen |
One lone pair, one double bond, up to one single bond |
N3t |
Nitrogen |
One lone pair, one triple bond |
N3b |
Nitrogen |
One lone pair, two aromatic bonds |
N5sc |
Nitrogen |
No lone pairs, up to four single bonds, charged +1 |
N5dc |
Nitrogen |
No lone pairs, one double bond, up to two single bonds, charged +1 |
N5ddc |
Nitrogen |
No lone pairs, two double bonds, charged +1 |
N5dddc |
Nitrogen |
No lone pairs, three double bonds, charged -1 |
N5tc |
Nitrogen |
No lone pairs, one triple bond, up to one single bond, charged +1 |
N5b |
Nitrogen |
No lone pairs, two aromatic bonds, up to one single bond |
O |
Oxygen |
No requirements |
Oa |
Oxygen |
Atomic oxygen with three lone pairs and no bonds |
O0sc |
Oxygen |
Three lone pairs, up to one single bond, charged -1 |
O0dc |
Oxygen |
Three lone pairs, one double bond, charged -2 |
O2s |
Oxygen |
Two lone pairs, up to two single bonds |
O2sc |
Oxygen |
Two lone pairs, up to one single bond, charged +1 |
O2d |
Oxygen |
Two lone pairs, one double bond |
O4sc |
Oxygen |
One lone pair, up to three single bonds, charged +1 |
O4dc |
Oxygen |
One lone pair, one double bond, up to one single bond, charged +1 |
O4tc |
Oxygen |
One lone pair, one triple bond, charged +1 |
Si |
Silicon |
No requirements |
Sis |
Silicon |
Up to four single bonds |
Sid |
Silicon |
One double bond (not to O), up to two single bonds |
SiO |
Silicon |
One double bond to an oxygen atom, up to two single bonds |
Sidd |
Silicon |
Two double bonds |
Sit |
Silicon |
One triple bond, up to one single bond |
Sib |
Silicon |
Two benzene bonds, up tp one single bond |
Sibf |
Silicon |
Three benzene bonds (fused aromatics) |
P |
Phosphorus |
No requirements |
P0sc |
Phosphorus |
Three lone pairs, up to one single bond, charged -2 |
P1s |
Phosphorus |
Two lone pairs, up to one single bond |
P1sc |
Phosphorus |
Two lone pairs, up to two single bonds, charged -1 |
P1dc |
Phosphorus |
Two lone pairs, one double bond, charged -1 |
P3s |
Phosphorus |
One lone pair, up to three single bonds |
P3d |
Phosphorus |
One lone pair, one double bond, up to one single bond |
P3t |
Phosphorus |
One lone pair, one triple bond |
P3b |
Phosphorus |
One lone pair, two aromatic bonds |
P5s |
Phosphorus |
No lone pairs, up to five single bonds |
P5sc |
Phosphorus |
No lone pairs, up to six single bonds, charged -1/+1/+2 |
P5d |
Phosphorus |
No lone pairs, one double bond, up to three single bonds |
P5dd |
Phosphorus |
No lone pairs, two double bonds, up to one single bond |
P5dc |
Phosphorus |
No lone pairs, one double bond, up to two single bonds, charged +1 |
P5ddc |
Phosphorus |
No lone pairs, two double bonds, charged +1 |
P5t |
Phosphorus |
No lone pairs, one triple bond, up to two single bonds |
P5td |
Phosphorus |
No lone pairs, one triple bond, one double bond |
P5tc |
Phosphorus |
No lone pairs, one triple bond, up to one single bond, charged +1 |
P5b |
Phosphorus |
No lone pairs, two aromatic bonds, up to one single bond, charged 0/+1 |
P5bd |
Phosphorus |
No lone pairs, two aromatic bonds, one double bond |
S |
Sulfur |
No requirements |
Sa |
Sulfur |
Atomic sulfur with three lone pairs and no bonds |
S0sc |
Sulfur |
Three lone pairs, up to once single bond, charged -1 |
S2s |
Sulfur |
Two lone pairs, up to two single bonds |
S2sc |
Sulfur |
Two lone pairs, up to three single bonds, charged -1/+1 |
S2d |
Sulfur |
Two lone pairs, one double bond |
S2dc |
Sulfur |
Two lone pairs, one to two double bonds, up to one single bond, charged -1 |
S2tc |
Sulfur |
Two lone pairs, one triple bond, charged -1 |
S4s |
Sulfur |
One lone pair, up to four single bonds |
S4sc |
Sulfur |
One lone pair, up to five single bonds, charged -1/+1 |
S4d |
Sulfur |
One lone pair, one double bond, up to two single bonds |
S4dd |
Sulfur |
One lone pair, two double bonds |
S4dc |
Sulfur |
One lone pair, one to three double bonds, up to three single bonds, charged -1/+1 |
S4b |
Sulfur |
One lone pair, two aromatic bonds |
S4t |
Sulfur |
One lone pair, one triple bond, up to one single bond |
S4tdc |
Sulfur |
One lone pair, one to two triple bonds, up to two double bonds, up to two single bonds, charged -1/+1 |
S6s |
Sulfur |
No lone pairs, up to six single bonds |
S6sc |
Sulfur |
No lone pairs, up to seven single bonds, charged -1/+1/+2 |
S6d |
Sulfur |
No lone pairs, one double bond, up to four single bonds |
S6dd |
Sulfur |
No lone pairs, two double bonds, up to two single bonds |
S6ddd |
Sulfur |
No lone pairs, up to three double bonds |
S6dc |
Sulfur |
No lone pairs, one to three double bonds, up to five single bonds, charged -1/-1/+2 |
S6t |
Sulfur |
No lone pairs, one triple bond, up to three single bonds |
S6td |
Sulfur |
No lone pairs, one triple bond, one double bond, up to one single bond |
S6tt |
Sulfur |
No lone pairs, two triple bonds |
S6tdc |
Sulfur |
No lone pairs, one to two triple bonds, up to two double bonds, up to four single bonds, charged -1/-1 |
Cl |
Chlorine |
No requirements |
Cl1s |
Chlorine |
Three lone pairs, zero to one single bonds |
Br |
Bromine |
No requirements |
Br1s |
Bromine |
Three lone pairs, zero to one single bonds |
I |
Iodine |
No requirements |
I1s |
Iodine |
Three lone pairs, zero to one single bonds |
F |
Fluorine |
No requirements |
F1s |
Fluorine |
Three lone pairs, zero to one single bonds |
He |
Helium |
No requirements, nonreactive |
Ne |
Neon |
No requirements, nonreactive |
Ar |
Argon |
No requirements, nonreactive |
Additionally, groups can also be defined as unions of other groups. For example,:
label="X_H_or_Xrad_H",
group=OR{X_H, Xrad_H},
12.1.2. Forbidden Groups¶
Forbidden groups can be defined to ban structures globally in RMG or to ban pathways in a specific kinetic family.
Globally forbidden structures will ban all reactions containing either reactants
or products that are forbidden. These groups are stored in in the file located at
RMG-database/input/forbiddenStructures.py.
To ban certain specific pathways in the kinetics
families, a forbidden group must be created, like the following group
in the intra_H_migration family:
forbidden(
label = "bridged56_1254",
group =
"""
1 *1 C 1 {2,S} {6,S}
2 *4 C 0 {1,S} {3,S} {7,S}
3 C 0 {2,S} {4,S}
4 *2 C 0 {3,S} {5,S} {8,S}
5 *5 C 0 {4,S} {6,S} {7,S}
6 C 0 {1,S} {5,S}
7 C 0 {2,S} {5,S}
8 *3 H 0 {4,S}
""",
shortDesc = u"""""",
longDesc =
u"""
""",
)
Forbidden groups should be placed inside the groups.py file located inside the
specific kinetics family’s folder RMG-database/input/kinetics/family_name/
alongside normal group entries. The starred atoms in the forbidden group
ban the specified reaction recipe from occurring in matched products and reactants.
In addition for forbidding groups, there is the option of forbidding specific
molecules or species in RMG-database/input/forbiddenStructures.py.
Forbidding a molecule will prevent that exact structure from being generated,
while forbidding a species will prevent any of its resonance structures from being
generated. Note that specific molecules or species can only be forbidden globally
and should not be specified in the groups.py file. To specify a forbidden molecule
or species, simply replace the group keyword with molecule or species:
# This forbids a molecule
forbidden(
label = "C_quintet",
molecule =
"""
multiplicity 5
1 C u4 p0
""",
shortDesc = u"""""",
longDesc =
u"""
""",
)
# This forbids a species
forbidden(
label = "C_quintet",
species =
"""
multiplicity 5
1 C u4 p0
""",
shortDesc = u"""""",
longDesc =
u"""
""",
)
12.1.3. Hierarchical Trees¶
Groups are ordered into the nodes of a hierarchical trees which is written at the end of groups.py. The root node of each tree is the most general group with the reacting atoms required for the family. Descending from the root node are more specific groups. Each child node is a subset of the parent node above it.
A simplified example of the trees for H-abstraction is shown below. The indented text shows the syntax in groups.py and a schematic is given underneath.
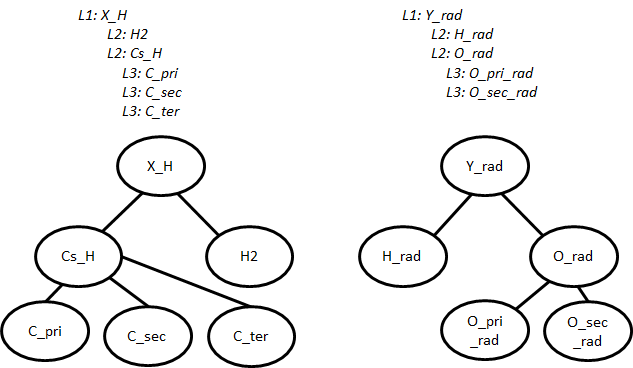
Individual groups only describe part of the reaction. To describe an entire reaction we need one group from each tree, which we call node templates or simply templates. (C_pri, O_pri_rad), (H2, O_sec_rad), and (X_H, Y_rad) are all valid examples of templates. Templates can be filled in with kinetic parameters from the training set or rules.
