2. Prune Edge Species¶
When dealing with complicated reaction systems, RMG calculation would easily hit the computer memory limitation. Memory profiling shows most memory especially during memory limitation stage is occupied by edge species. However, most edge species in fact wouldn’t be included in the core (or final model). Thus, it’s natural to get rid of some not “so useful” edge species during calculation in order to achieve both low memory consumption and mechanism accuracy. Pruning is such a way.
2.1. Key Parameters in Pruning¶
toleranceKeepInEdgeAny edge species to prune should have peak flux along the whole conversion course lower than
toleranceKeepInEdge\(*\) characteristic flux. Thus, larger values will lead to smaller edge mechanisms.toleranceMoveToCoreAny edge species to enter core model should have flux at some point larger than
toleranceMoveToCore\(*\) characteristic flux. Thus, in general, smaller values will lead to larger core mechanisms.toleranceInterruptSimulationOnce flux of any edge species exceeds
toleranceInterruptSimulation\(*\) characteristic flux, dynamic simulation will be stopped. Usually this tolerance will be set a very high value so that any flux’s exceeding that means mechanism is too incomplete to continue dynamic simulation.maximumEdgeSpeciesIf dynamic simulation isn’t interrupted in half way and total number of the edge species whose peak fluxes are higher than
toleranceKeepInEdge\(*\) characteristic flux exceedsmaximumEdgeSpecies, such excessive amount of edge species with lowest peak fluxes will be pruned.minCoreSizeForPruneEnsures that a minimum number of species are in the core before pruning occurs, in order to avoid pruning the model when it is far away from completeness. The default value is set to 50 species.
minSpeciesExistIterationsForPruneSet the number of iterations an edge species must stay in the job before it can be pruned. The default value is 2 iterations.
2.2. How Pruning Works¶
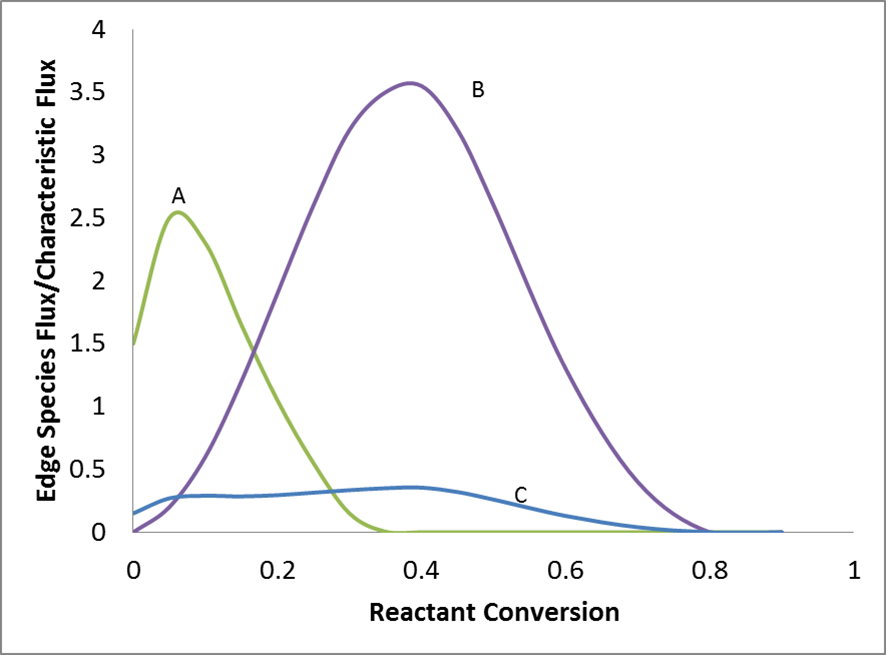
The goal of pruning is to delete those “useless” edge species. So “usefulness” should be defined and it’s natural to have flux as a
criterion for “usefulness”. Since flux changes with reactant conversion, peak flux is chosen here to make decision of pruning or not.
Every time pruning is triggered, edge species with peak flux lower than toleranceKeepInEdge \(*\) characteristic flux will be deleted.¶
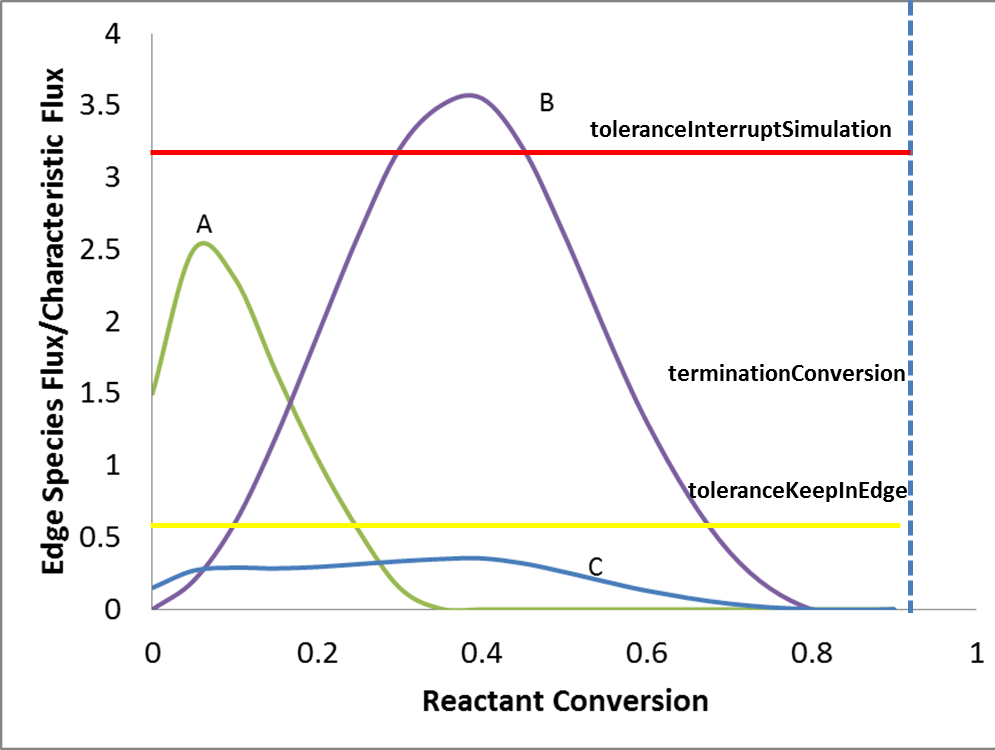
However, pruning is not always triggered because of toleranceInterruptSimulation. As mentioned above, in order to prune, RMG needs to figure out
the peak flux of each edge species, which requires dynamic simulation to complete. If some run of dynamic simulation is terminated in half way
by toleranceInterruptSimulation, pruning is rejected although there might be some edge species with peak fluxes lower than
toleranceKeepInEdge \(*\) characteristic flux. Since pruning requires to complete dynamic simulation, setting toleranceInterruptSimulation to be positive infinity,
as an extreme case, means always enabling pruning. Another extreme case would be that it has same value as toleranceMoveToCore where
no pruning occurs.¶
In summary, each run of dynamic simulation will proceed towards terminationConversion unless some flux exceeds
toleranceInterruptSimulation \(*\) characteristic flux. Following complete simulation is the pruning of edge species whose flux is not high enough be kept
in the edge, which is followed by pruning of excessive amount of edge species to make sure total edge species number is no greater than maximumEdgeSpecies.
